Abstract
Approximately more than half of the high score serious cancers of epithelial ovarian suffers the changes in the gene of homologous recombination, while the remaining make changes in other repairs of DNA ways at excessive rate or frequency (Somyajit et al., 2010). Such gene alteration that are cancer specific can consult selective amount of responsiveness to harmful agents of DNA like carboplatin and cisplatin, etoposide, topotecan, doxorubicin and gemcitabine. PARP inhibitors that in the near past have been frequently used by the FDA in order to treat the advanced cancer caused in the ovaries. This also work as DNA destructive agent by prompting complexes of PARP-DNA. The DNA destructive agents make many other kinds of DNA that need specific DNA in order to repair DNA genes. This usually convince duplication fork slowing, it is also known as stress. Duplication of stress generally produce or triggers DNA repairs in the form of checkpoints proteins, these proteins further prevent the DNA damaging. Therefore, the targeting of DNA repair genes or the checkpoints of DNA repair argument the anti-tumor activity of agents responsible for DNA injuring. The research described the balanced approach to use the DNA repair and checkpoint of DNA repairs acts as a sole agent. This researchhas also presented the strategies for combining these agents with damaging DNA agents of the therapy of ovarian cancer based on some specific genes alteration. There is accruing indication that it may certainly be achieved, and that cellular stress can be minimized by cure with select pharmacological agents. The ethics for this new approach to cure cancer, as well as representative ER stress-aggravating compounds, will be presented in this review.
Study Aim
The aim of this study is to compare the effects of anti-cancer drug Olaparib in two human cancer cell lines with normal and aberrant expression of the DNA repair gene BRCA1.
Study Objectives
The objectives of this study include:
- To gain experience in working with human tissue culture, which includes:
- Handling of adherent cancer cell lines;
- Maintaining stable cell lines over-expressing DNA repair proteins of the interest.
- Asses the molecular effect of the anti-cancer drugs, Olaparib and Cisplatin, on:
- Genome stability;
- Cell proliferation;
- Cell apoptosis.
- Analyze chromosomal composition in cells with different BRCA status after treatment with anticancer drugs Olaparib and Cisplatin.
Introduction
The maintenance of genome integrity is the most important commission in the life of cell or entire organism. The integrity on the chromosomal or gene level allows normal functioning and survival. Any DNA lesions lead to the cell cycle arrest, followed by either successful DNA repair or mutation-triggered apoptosis.
However, a specific set of genetic mutations permits to avoid cell cycle arrest and overcome all check points leading to uncontrollable cellular growth and resulting in cancerous transformation.
Cancer became a leading life-threatening disease in the modern world. According to the GLOBOCAN study, over 14 million new cancer cases and over 8 million cancer related deaths were registered in 2012 worldwide (Ferlay et al., 2015). Currently, 27 major types of cancer described. Among those, lung cancers account for 1.82 million, breast cancers for 1.67 million and colorectal cancers for 1.36 million of commonly diagnosed cases (Ferlay et al., 2015). Lung cancers are the main type of cancers linked to mortality. The treatment of breast cancers have advanced in recent year, which allows to significantly extend disease-free survival.
Continuous cellular stress is a major basis of deteriorating genome integrity, which compromises extracellular environment and cause damage to DNA, RNA and proteins structure and functions (Poljsak & Milisav, 2012). There are many stressors, and different kinds of them infer different cellular responses, impairments and repair mechanisms. Most of the stressors are triggering damages to DNA structure. Persistent stress generates mutations and, as a result, increases susceptibility to cancer and ageing (Poljsak & Milisav, 2012). Excessive stress-related damage results in the induction of autophagy and apoptosis (Poljsak & Milisav, 2012).
Type of mutations and DNA repair machinery
DNA damage is generally classified by its source as oxidative, chemical, hydrolytic and radiation induced. The type of damage has corresponding structural features such as deamination of bases, base conversions, formations of DNA-adducts, and DNA strand breaks. Each type of DNA damage requires its own mechanism for the repair. Disturbances in DNA repair pathways can predispose cells to accumulating damage-specific DNA mutations.
The most dangerous DNA injuries are single- and double-strand breaks of DNA chain. They are recognized as main drivers of carcinogenic processes (Aparicio, Baer, & Gautier, 2014). Single strand break (SSB), if left unrepaired, can invade the intact double helix, insert single-stranded chain of DNA into it and create a triple helix structure. This often results in the formation of DNA double strand break (DSB). As opposed to most of the types of DNA damage that could be repaired with little homology required, DNA strand breaks can be repaired by homologous recombination (HR) without sequence changes only with a large homological DNA stretch used as a template. This repair can be performed only at specific cell phase, S phase, when sister chromatids are present and provide that homology. At the G1, G2 or G0 cell cycle phases the repair also could be executed but mainly by non-homologous end joining (NHEJ) which does not require extensive homology and is therefore prone to errors (Kelley, Logsdon, & Fishel, 2014).
DNA strand breaks and its repair
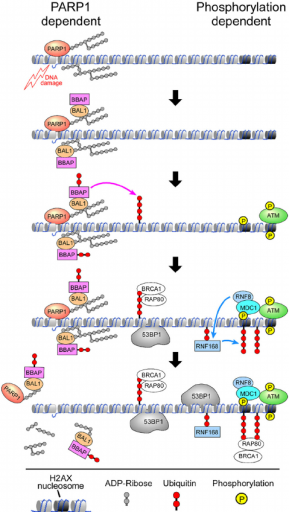
The scheme of DNA repair pathway driven by homologous recombination is shown in Fig. 1. As found previously, PARP activation is the earliest response to a single or double strand DNA break. The protein PARP facilitates several enzymatic reactions at the site of DNA damage keeping the broken ends together and relaxing the surrounding chromatin, thus making the site available for the repair protein complexes (Yan et al., 2013). Following PARP activation, the ATM protein is recruited to the site. This protein determines which repair pathway will be activated: NHEJ or HR (Aparicio et al., 2014). There is not clear understanding about the mechanism of pathway choice. However, it is known that HR requires activity of BRCA 1 and 2 proteins (Zhang & Powell, 2005), as well as activity of PCNA (Hasan, Hassa, Imhof, & Hottiger, 2001).
Figure 1. Schematic view of homologous recombination of DNA strand breaks (Yan et al., 2013)
BRCA1
BRCA1 gene is mapped on the chromosome 17q21. The gene encodes a 1,863 amino acid protein containing several zinc-finger domains close to the N terminus of the protein. High level of BRCA1 expression is found in breast, ovary and testes tissues.
Mutations in BRCA1 and BRCA2 genes are often detected in familial breast and ovarian cancers. BRCA1 mutations are prevalent in hereditary breast cancer accounting for 45 percent of clinically relevant findings, and BRCA1 mutations are even more prevalent in the families with hereditary breast and ovarian cancers, together accounting for 80 percent of the cases (Zhang & Powell, 2005). Mutations in BRCA1 gene are associated with breast and ovarian cancers and have multifactorial or autosomal dominant inheritance.
In addition, the BRCA1 protein expression is significantly compromised or absent in sporadic breast malignancies (Zhang & Powell, 2005). Cellular function of BRCA1 protein is described as a critical component in DNA repair and cell cycle check point control (Wang et al., 2009). Another direct function of BRCA1 protein is a modulation of chromatin structure, namely chromatin decondensation, which is required for the control of transcription (Bochar et al., 2000; Ye et al., 2001).
PARP1
The poly(ADP-ribose) polymerase (PARP) is a gene mapped on chromosome 1q42.12. The protein is nuclear and co-localized with nucleosomal core histones. The enzyme catalyzes the transfer of ADP-ribose to a number of nuclear proteins. The activity of enzyme is induced by the presence of DNA single strand breaks, which provide substrate for the PARP enzymatic activity (Alkhatib et al., 1987). ADP-ribosylation is a DNA break dependent protein modification, which serves as a signal for DNA repair needs. There is a positive correlation between PARP activity level of individuals and their life span, which probably reflects the efficacy of the genome integrity maintenance (Grube & Burkle, 1992). Immunostaining showed that PARP proteins are co-localized with mitotic spindle, and hydrolytic breakage of the protein resulted in rapid spindle disassembly, which blocked the correct chromosomal segregation during cell division (Chang, Jacobson, & Mitchison, 2004). PARP is required for gamma-H2AX and Rad51 foci formation at the site of DNA break, triggering DNA repair and recruitment of ATM to the site of DNA damage (Bryant et al., 2005). The absence of PARP at the DNA break site or its inhibition causes collapse of replication forks via the broken DNA end intervention arresting the cells in S phase (Bryant et al., 2005). Moreover, BRCA deficient cells are very sensitive to the PARP enzymatic activity levels. Impaired function of PARP in BRCA deficient cells leads to the cycle arrest and cell death (Bryant et al., 2005; Farmer et al., 2005).
DNA repair pathways in targeted anticancer therapy
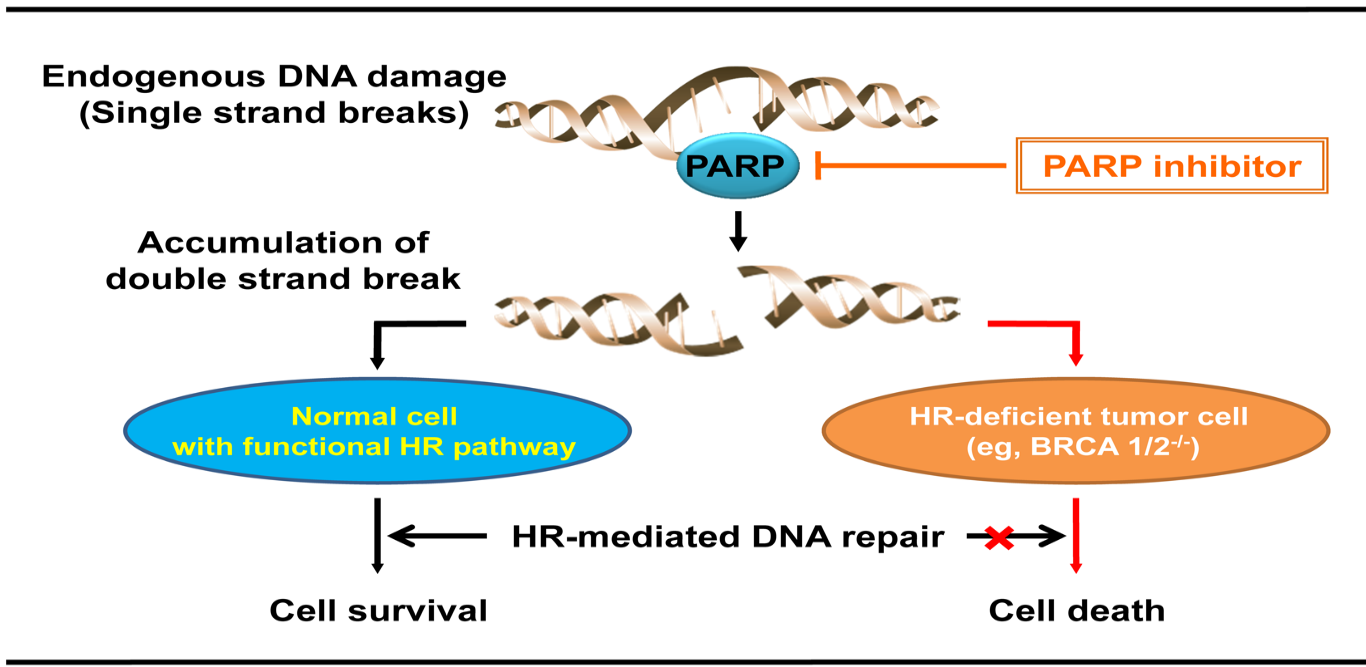
The observed effect of PARP inhibition on apoptosis in the BRCA deficient cells led to the idea of targeted anticancer therapy (Fig. 2). Monotherapy by PARP inhibitors could be used to treat BRCA deficient tumors (Bryant et al., 2005; Farmer et al., 2005; Fong et al., 2009). Such approach ensures lower toxicity for the patients. The improved toxicity profile originates from the fact that only cells with the lost BRCA activity are subjects to the PARP inhibitor-induced cell death (Fong et al., 2009). The mechanism of PARP-targeted antitumor therapy consists of restricting the DNA break repair in the cells already deficient in homologous recombination due to BRCA dysfunction. This leads to the accumulation of DNA damage, cell cycle arrest and cell death triggered by these factors (Helleday, 2011). As demonstrated in vitro and by using xenografts, BRCA aberrant cells are drastically more sensitive to the PARP inhibitors compared to the normal cell (Fong et al., 2009).
Figure 2. PARP-targeted cancer therapy in BRCA deficient cells
Currently, several PARP inhibitors including olaparib, veliparib, niraparib, rucaparib and talazoparib are tested for targeted anticancer therapy.
Olaparib
Nine clinical trials were conducted to prove safety and efficacy of PARP targeted therapy in BRCA deficient breast cancers (Javle & Curtin, 2011). The most promising results were obtained with Olaparib. A phase 1 clinical trial on breast cancer patients carrying BRCA mutation demonstrated that Olaparib provides clinical benefits in 63 percent of cases, with positive tumor marker responses and significant disease stabilization (Fong et al., 2009). In a phase 2 clinical trial on the BRCA deficient ovarian cancer patients, efficacy and safety of Olaparib was also confirmed (Konecny & Kristeleit, 2016).
Cisplatin
Cisplatin is another widely used compound for anticancer chemotherapy. In many cases, patients develop reduced sensitivity to cisplatin (or even have it at the start). The latest investigation showed that synergetic use of PARP inhibitor olaparib sensitizes cancer cells to cisplatin in cervical cancer patients (Wang et al., 2017). Cisplatin toxicity could be overcome via PARP1 regulation. This will possibly provide future direction for PARP based approaches beyond the BRCA deficient cancers (Singh, Chauhan, & Kang, 2018; Xu et al., 2017).
Material and Methods
Cell lines
In the present study, two cancer cell lines were used: MCF7 cells derived from adenocarcinoma of breast, and HeLa Kyoto cells genetically modified to stably express the fused protein green fluorescent protein-tagged proliferating cell nuclear antigen (GFP-PCNA). Both cell lines have epithelial origin. However, the cell lines derive from different cancerous tissues: MCF7was obtained from metastatic side of breast cancer patient, and HeLa was established from cervical cancer specimen. The cell lines differ in BRCA1 status.
MCF7 is characterized by loss of heterozygosity at BRCA1 locus, which is reflected in reduction of BRCA1 expression level (Elstrodt et al., 2006; Kachhap et al., 2001; Vissac et al., 2002). The karyotype of MCF7 cells has modal chromosome number equal to 82 on average, ranging from 66 to 87 chromosomes per nucleus, which corresponds to hypertriploidy or hypotetraploidy. The cells carry multiple marker chromosomes, up to 34 marker chromosomes per cell. Approximately, one third of cellular population has markers in their nuclei. In the vast majority of cells with the marker chromosomes, those chromosomes are present as a single large submetacentric marker chromosome, three large subtelocentric marker chromosomes and a set of smaller marker chromosomes (Rondon-Lagos et al., 2014). Chromosome 20 is deleted. Despite pronounced transformation and chromosomal abnormality, MCF7 cells still feature characteristics of differentiated mammary epithelium. MCF7 cells are estrogen sensitive and express WNT oncogene.
In contrast, HeLa Kyoto cell line carries two normal, not mutated BRCA1 genes that have not changed their expression levels compared to healthy cervical epithelium (Landry et al., 2013; Vissac et al., 2002). The karyotype of HeLa cells shows multiple rearrangements and changed ploidy with modal chromosome number of 82, ranging from 70 to 164 chromosomes per cell. As shown by G-banding, almost a hundred percent of cells carry multiple marker chromosomes originated from the fusion between chromosomes 1 and 3, 3 and 5, and 11 and 19 (Chen, 1988).
The GFP-Flag-PCNA carrying HeLa-Kyoto cells are a great instrument to study cell cycle by microscopy. Previous studies demonstrated that the PCNA gene expression is clearly associated with S-phase of cell cycle. It was previously shown that PCNA expression marks the beginning of S-phase, and the product becomes undetectable shortly before the cells enter the next phase of the cell cycle (Celis et al., 1986; Kurki et al., 1986).
DNA-damaging reagents
In the present study, two anticancer drugs acting via PARP1 were used: Olaparib and Cisplatin (Sigma-Aldrich, USA).
Olaparib (SIGMA-Aldrich)
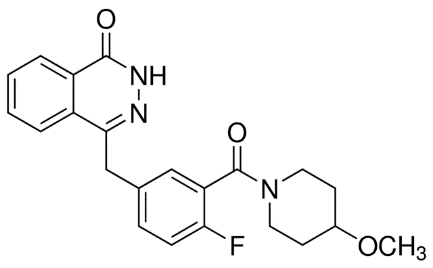
Olaparib is the name for 4-[4-Fluoro-3-[(4-methoxypiperidin-1-yl) carbonyl] benzyl] phthalazin-1(2H)-1, the organic compound with molecular weight of 395.43 Da and empirical formula C22H22FN3O3 (Fig. 3).
Figure 3. Molecular structure of Olaparib.
Olaparib is a PARP inhibitor, which induces lethality in BRCA1/2 deficient cells carrying either homologous deleterious BRCA mutations or compound heterozygotes for those genes (Audeh et al., 2010). Olaparibe demonstrates anti-proliferative effect in cells with BRCA1 and BRCA2 mutations and cells with BRCA1 allelic loss. In addition, there is an evidence of autophagy/mitophagy induction by Olaparib in BRCA deficient cells (Arun et al., 2015).
Cisplatin (SIGMA-Aldrich)
Cisplatin is the name for cis-Diammineplatinum(II) dichloride, a chemical with molecular weight of 300.05 Da and linear formula Pt(NH3)2Cl2. PARP1 is one of the targets for cisplatin and its prodrugs (Xu et al., 2017). Cisplatin reacts with water and DNA molecules and forms bulky adducts with DNA, thus preventing replication. The consequent interaction of cisplatin and DNA complex with PARP1 protein results in cell death (Michels et al., 2013).
Cell culture
The cells were grown in DMEM media containing 10% FCS, supplied with L-glutamine and antibiotics according to the ACCT repository instructions. The cells were split twice per week in 1 to 5 ratio using trypsinization procedure (Surendranath, 2014)
Cell proliferation and viability test
To determine cell proliferation and viability, Resuzurin Viability Kit (Biotium Inc., USA) was used. All steps were performed according to the manufacturer’s protocol. The date were obtained by measuring the absorbance at 570 nm and 600 nm using an absorbance microplate reader for colorimetric detection, and by measuring fluorescence with excitation/emission at 570/585 nm using a fluorescence microplate reader for fluorescent based detection. Absorbance values were adjusted to background level and plotted against the density of plated cell. The optimal values were identified for future use.
Metaphase Spreads Preparation:
Metaphase spreads were prepared using standard Colcemid (Life Technologies) arrest procedure followed by hypotonic treatment and standard methanol/acetic acid fixation procedures (Surendranath, 2015)
Olaparib and Cisplatin treatment
To study the effect of Olaparib and Cisplatin, MCF7 and HeLa Kyoto cells were plated on the cover slips placed into 6 well culture plates or directly into 12 well culture plates and grown to 70% confluence. The culture media was removed, cells were washed three times with 1XPBS to remove any traces of FCS and then DMEM with 0.2% FCS was added for 16 hours to synchronize the cells. Synchronization level of cell cultures was assessed by live fluorescent microscopy detecting the green fluorescence produced by GFP-PCNA fusion construct product in HeLa Kyoto cells.
After the starvation step, low serum media was replaced with complete DMEM media containing 10% FCS and supplemented with either Olaparib or Cisplatin at different concentrations. Olaparib concentrations were 0, 100, 200 and 300 nM, and Cisplatin concentrations were 0, 0.15, 0.3, 0.6, 1.25, 2.5, 5, 10, 20, 30 and 40 mM. The cells were incubated at 370C for 48 hours and then subjected to live microscopy, cell proliferation assay, cell viability assay or Giemsa staining.
To perform this procedure, compare the effects of the definite anti-cancer drug Olaparib in two separate human ovarian cancer cell lines, in the BRCA wild type and in the BRCA mutant. Following are the procedures that were applied
• Genome stability
• Cell proliferation
• Cell apoptosis
Results
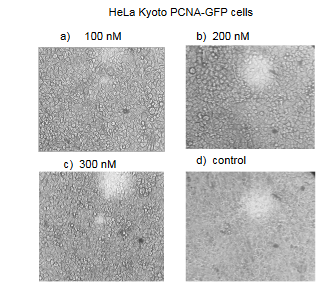
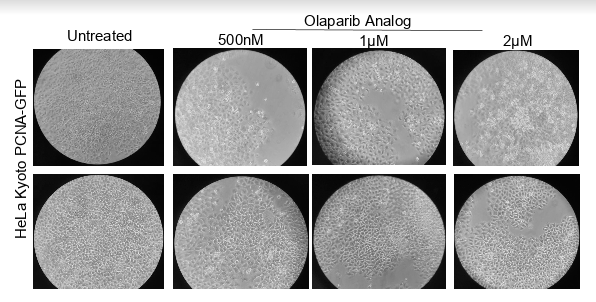
Figure: Images of HeLa Kyoto GFP-PCNA cells treated for 48 hours with different concentrations of Olaparib.
Figure : Images of HeLa Kyoto GFP-PCNA cell in visible and fluorescent light.
Figure: Hela Kyoto PCNA-GFP were plated in multi- well dishes and treated with Olaparib with increasing concentration for 48 hours period and viewed under bright field microscope and further these were fixed and resazurin was added.
Discussion
It is to visualize the duplication or replication in the cells. The CHO cells were generated that stably express human PCNA which is tagged with GFP. Some other test expressed that GFP-hPCNA with the anti PCNA antibodies showed it that the level of expression was similar.
Duplication of the genome of mammals starts at several thousands of origins that are generally turned on at diverse time during its phase. However, a specific set of genetic mutations permits to avoid cell cycle arrest and overcome all check points leading to uncontrollable cellular growth and resulting in cancerous transformation.
Induced DNA duplication stress is the key to instability of chromosome in caner to human. This is supposed to play the role in the formation of oncogenesis. It is imperative, hence we not only try to understand the cause of stress, but we also try to cover the path ways that are used by the cells to show its potential effects oncogenically. When it is challenged by the suplication stress, the program of duplication that is taking place at CFSs is often assigned to such an extent that it continues to synthesis DNA even after the point where cell has managed to enter to the initial phase of mitosis. The recent researches have found that it is not conventional to synthesis DNA.
References
Alkhatib, H. M., Chen, D. F., Cherney, B., Bhatia, K., Notario, V., Giri, C., . . . Smulson, M. E. (1987). Cloning and expression of cDNA for human poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A, 84(5), 1224-1228.
Aparicio, T., Baer, R., & Gautier, J. (2014). DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst), 19, 169-175. doi:10.1016/j.dnarep.2014.03.014
Arun, B., Akar, U., Gutierrez-Barrera, A. M., Hortobagyi, G. N., & Ozpolat, B. (2015). The PARP inhibitor AZD2281 (Olaparib) induces autophagy/mitophagy in BRCA1 and BRCA2 mutant breast cancer cells. Int J Oncol, 47(1), 262-268. doi:10.3892/ijo.2015.3003
Audeh, M. W., Carmichael, J., Penson, R. T., Friedlander, M., Powell, B., Bell-McGuinn, K. M., . . . Tutt, A. (2010). Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet, 376(9737), 245-251. doi:10.1016/S0140-6736(10)60893-8
Bochar, D. A., Wang, L., Beniya, H., Kinev, A., Xue, Y., Lane, W. S., . . . Shiekhattar, R. (2000). BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell, 102(2), 257-265.
Bryant, H. E., Schultz, N., Thomas, H. D., Parker, K. M., Flower, D., Lopez, E., . . . Helleday, T. (2005). Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature, 434(7035), 913-917. doi:10.1038/nature03443
Celis, J. E., Madsen, P., Nielsen, S., & Celis, A. (1986). Nuclear patterns of cyclin (PCNA) antigen distribution subdivide S-phase in cultured cells–some applications of PCNA antibodies. Leuk Res, 10(3), 237-249.
Chang, P., Jacobson, M. K., & Mitchison, T. J. (2004). Poly(ADP-ribose) is required for spindle assembly and structure. Nature, 432(7017), 645-649. doi:10.1038/nature03061
Chen, T. R. (1988). Re-evaluation of HeLa, HeLa S3, and HEp-2 karyotypes. Cytogenet Cell Genet, 48(1), 19-24. doi:10.1159/000132579
Dasari, S. and Tchounwou, P.B., 2014. Cisplatin in cancer therapy: molecular mechanisms of action. European journal of pharmacology, 740, pp.364-378.
Elstrodt, F., Hollestelle, A., Nagel, J. H., Gorin, M., Wasielewski, M., van den Ouweland, A., . . . Schutte, M. (2006). BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res, 66(1), 41-45. doi:10.1158/0008-5472.CAN-05-2853
Farmer, H., McCabe, N., Lord, C. J., Tutt, A. N., Johnson, D. A., Richardson, T. B., . . . Ashworth, A. (2005). Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature, 434(7035), 917-921. doi:10.1038/nature03445
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., . . . Bray, F. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer, 136(5), E359-386. doi:10.1002/ijc.29210
Fong, P. C., Boss, D. S., Yap, T. A., Tutt, A., Wu, P., Mergui-Roelvink, M., . . . de Bono, J. S. (2009). Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med, 361(2), 123-134. doi:10.1056/NEJMoa0900212
Grube, K., & Burkle, A. (1992). Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc Natl Acad Sci U S A, 89(24), 11759-11763.
Hasan, S., Hassa, P. O., Imhof, R., & Hottiger, M. O. (2001). Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature, 410(6826), 387-391. doi:10.1038/35066610
Helleday, T., Petermann, E., Lundin, C., Hodgson, B. and Sharma, R.A., 2008. DNA repair pathways as targets for cancer therapy. Nature Reviews Cancer, 8(3), p.193.
Helleday, T. (2011). The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Molecular Oncology, 5(4), 387-393. doi:https://doi.org/10.1016/j.molonc.2011.07.001
Javle, M., & Curtin, N. J. (2011). The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer, 105(8), 1114-1122. doi:10.1038/bjc.2011.382
Kachhap, S. K., Vetale, S. P., Dange, P., & Ghosh, S. N. (2001). Reduced expression of the BRCA1 gene and increased chromosomal instability in MCF-7 cell line. Cell Biol Int, 25(6), 547-551. doi:10.1006/cbir.2000.0657
Kelley, M. R., Logsdon, D., & Fishel, M. L. (2014). Targeting DNA repair pathways for cancer treatment: what’s new? Future Oncol, 10(7), 1215-1237. doi:10.2217/fon.14.60
Konecny, G. E., & Kristeleit, R. S. (2016). PARP inhibitors for BRCA1/2-mutated and sporadic ovarian cancer: current practice and future directions. Br J Cancer, 115(10), 1157-1173. doi:10.1038/bjc.2016.311
Kurki, P., Vanderlaan, M., Dolbeare, F., Gray, J., & Tan, E. M. (1986). Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle. Exp Cell Res, 166(1), 209-219.
Landry, J. J., Pyl, P. T., Rausch, T., Zichner, T., Tekkedil, M. M., Stutz, A. M., . . . Steinmetz, L. M. (2013). The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda), 3(8), 1213-1224. doi:10.1534/g3.113.005777
Levran, O., Attwooll, C., Henry, R.T., Milton, K.L., Neveling, K., Rio, P., Batish, S.D., Kalb, R., Velleuer, E., Barral, S. and Ott, J., 2005. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nature genetics, 37(9), p.931.
Meindl, A., Hellebrand, H., Wiek, C., Erven, V., Wappenschmidt, B., Niederacher, D., Freund, M., Lichtner, P., Hartmann, L., Schaal, H. and Ramser, J., 2010. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nature genetics, 42(5), p.410.
Michels, J., Vitale, I., Senovilla, L., Enot, D. P., Garcia, P., Lissa, D., . . . Kroemer, G. (2013). Synergistic interaction between cisplatin and PARP inhibitors in non-small cell lung cancer. Cell Cycle, 12(6), 877-883. doi:10.4161/cc.24034
Minami, D., Takigawa, N., Takeda, H., Takata, M., Ochi, N., Ichihara, E., Hisamoto, A., Hotta, K., Tanimoto, M. and Kiura, K., 2013. Synergistic effect of olaparib with combination of cisplatin on PTEN-deficient lung cancer cells. Molecular Cancer Research, 11(2), pp.140-148.
Özer, Ö., Bhowmick, R., Liu, Y. and Hickson, I.D., 2018. Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget, 9(22), pp.15836-15846.
Poljsak, B., & Milisav, I. (2012). Clinical implications of cellular stress responses. Bosn J Basic Med Sci, 12(2), 122-126. doi:10.17305/bjbms.2012.2510
Rondon-Lagos, M., Verdun Di Cantogno, L., Marchio, C., Rangel, N., Payan-Gomez, C., Gugliotta, P., . . . Sapino, A. (2014). Differences and homologies of chromosomal alterations within and between breast cancer cell lines: a clustering analysis. Mol Cytogenet, 7(1), 8. doi:10.1186/1755-8166-7-8
Schlacher, K., Wu, H. and Jasin, M., 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer cell, 22(1), pp.106-116.
Singh, M. P., Chauhan, A. K., & Kang, S. C. (2018). Morin hydrate ameliorates cisplatin-induced ER stress, inflammation and autophagy in HEK-293 cells and mice kidney via PARP-1 regulation. Int Immunopharmacol, 56, 156-167. doi:10.1016/j.intimp.2018.01.031
Somyajit, K., Subramanya, S. and Nagaraju, G., 2010. RAD51C: a novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer. Carcinogenesis, 31(12), pp.2031-2038.
Vissac, C., Peffault De Latour, M., Communal, Y., Bignon, Y. J., & Bernard-Gallon, D. J. (2002). Expression of BRCA1 and BRCA2 in different tumor cell lines with various growth status. Clin Chim Acta, 320(1-2), 101-110.
Wang, B., Hurov, K., Hofmann, K., & Elledge, S. J. (2009). NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev, 23(6), 729-739. doi:10.1101/gad.1770309
Wang, J., Kho, D. H., Zhou, J. Y., Davis, R. J., & Wu, G. S. (2017). MKP-1 suppresses PARP-1 degradation to mediate cisplatin resistance. Oncogene, 36(43), 5939-5947. doi:10.1038/onc.2017.197
Xu, Z., Hu, W., Wang, Z., & Gou, S. (2017). Platinum(IV) prodrugs multiply targeting genomic DNA, histone deacetylases and PARP-1. Eur J Med Chem, 141, 211-220. doi:10.1016/j.ejmech.2017.09.074
Yan, Q., Xu, R., Zhu, L., Cheng, X., Wang, Z., Manis, J., & Shipp, M. A. (2013). BAL1 and its partner E3 ligase, BBAP, link Poly(ADP-ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8. Mol Cell Biol, 33(4), 845-857. doi:10.1128/MCB.00990-12
Ye, Q., Hu, Y. F., Zhong, H., Nye, A. C., Belmont, A. S., & Li, R. (2001). BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J Cell Biol, 155(6), 911-921. doi:10.1083/jcb.200108049
Zhang, J., & Powell, S. N. (2005). The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res, 3(10), 531-539. doi:10.1158/1541-7786.MCR-05-0192