Introduction
Salting out is one of the methods used to extract DNA material of high quality within the shortest time in the laboratory. Researchers have developed several other methods to ensure the adequate availability of high-quality DNA for various molecular research purposes. Even though the procedures vary considerably, they involve three main steps: lysis of the cell wall, cell membrane, and nuclear membrane, isolation of DNA from other cell inclusions, and purification of the extracted DNA. All the methods employ agarose gel electrophoresis to separate the DNA molecules. The experiment aimed at extracting DNA from kiwi fruit and strawberry fruit, and comparing the salting out approach to the YouTube method of extraction.
DNA is life’s blueprint, and it is found in all organisms, from viruses to human beings. It is the hereditary material that transmits traits from one generation to the other. Moreover, DNA carries the template that the body uses for protein synthesis. Behavior, personality, body size, fingerprints, and skin color are all determined by the blueprint encoded in the DNA. For instance, the difference between races is a result of variation in DNA content. Additionally, some diseases, such as diabetes mellitus, sickle cell anemia, albinism, and Huntington’s disease, are all passed from the parents to their children through DNA. That is why these diseases are more prevalent in people who come from families where some members have suffered the same illness. In plants, the DNA determines the maturity rate, the size of fruits, and fruit color. The plant DNA also determines drought resistance and disease resistance.
DNA extraction isolates DNA strands from the cells of living organisms. Given the momentousness of the DNA, it is one area that has adequately intrigued many researchers. Friedrich Miescher discovered DNA extraction in 1869 (Tamari and Hinkley, 2016 p240), and since that time, researchers across the ages have employed techniques to isolate DNA materials from plants and animals for research purposes. For instance, DNA extraction has been used in the production of genetically modified organisms (GMOs) pioneered by Harbert Boyer and Stanley Cohen in 1973 (Zhang, Sun, and Ma, 2017 p412). DNA extraction has also been used in forensic medicine and criminology to prove the identity of suspects of such crimes as rape, murder, and robbery. Doctors also depend on DNA extraction for differential diagnosis and treatment of various diseases.
Method
Equipment and Reagents Used
- Kiwi and Strawberry fruit tissues
- Proteinase K
- NES buffer solution
- Incubator
- Auto pipette
- Centrifuge
- Micro-centrifuge tubes
- Razor blade
- Ethanol 70%
- Ethanol 100%
- 5M NaCl
- Agarose Gel
- Loading dye
Procedure Followed
Samples were labeled with the fruit types as K for S for Kiwi and Strawberry, respectively. The samples had been prepared earlier in the following manner: Fruit tissues were finely chopped using a razor blade and then placed in 1.5mL micro-centrifuge tubes. To break down the proteins in the fruit tissues, ten microliters of proteinase K were then added into the tubes and shaken gently. After that, 600 µL of TNES buffer was added to maintain a stable pH which is ideal for the survival of the fruit tissues. Additionally, the buffer solution aided cell lysis. The contents of the microcentrifuge tubes were then incubated overnight at 37oC.
Into each sample, one hundred and seventy microliters of 5M NaCl were added and flicked for ten seconds to mix. The purpose of 5M NaCl was to precipitate proteins from the sample. The microcentrifuge tubes and the contents thereof were then micro-centrifuged for 5 minutes at 12000 rpm to separate the precipitated proteins from the other contents of the microcentrifuge tubes.
Other clean micro-centrifuge tubes were labeled, and into each tube was added 300 µL of the supernatant. The supernatant was the liquid part that contained the DNA material. Three hundred microliters of 100% ethanol were then auto-pipetted and added to the supernatant in the new tube. The tube was then inverted three times gently to mix the contents. The purpose of adding 100% ethanol was to precipitate DNA. The mixture was then incubated for 10 minutes at room temperature. After incubation, the tubes were micro-centrifuged for 5 minutes at 12000 rpm so that the precipitated DNA could settle at the bottom.
The supernatant was then carefully removed using an autopipette. Any remaining traces of ethanol were completely drained by placing the tube upside down. The remaining DNA pellets were then washed with 300 µL of cold 70% ethanol to remove any lingering protein materials. The tube was again micro centrifuged for five minutes at 12000 rpm to ensure a complete pill of the DNA. The 70% ethanol was then discarded using a micropipette. The microcentrifuge tube was then opened and placed on a heat block set at 30C for twenty minutes to ensure that all the ethanol evaporated. Seeker et al. (2016 p137) assert that lingering traces of ethanol would prevent the dissolution of DNA and also impede the process of gel electrophoresis. Therefore all the ethanol has to be evaporated completely.
The DNA remaining in the tube was then dissolved by adding 20 µL of water into the tube and mixing gently. To a clean tube, 2 µL of the loading dye was put in, followed by 10 µL of the sample solution. The mixture of sample and loading dye was then loaded onto the agarose gel. The gel was run, and the image obtained is shown in figure 1.
Results Obtained
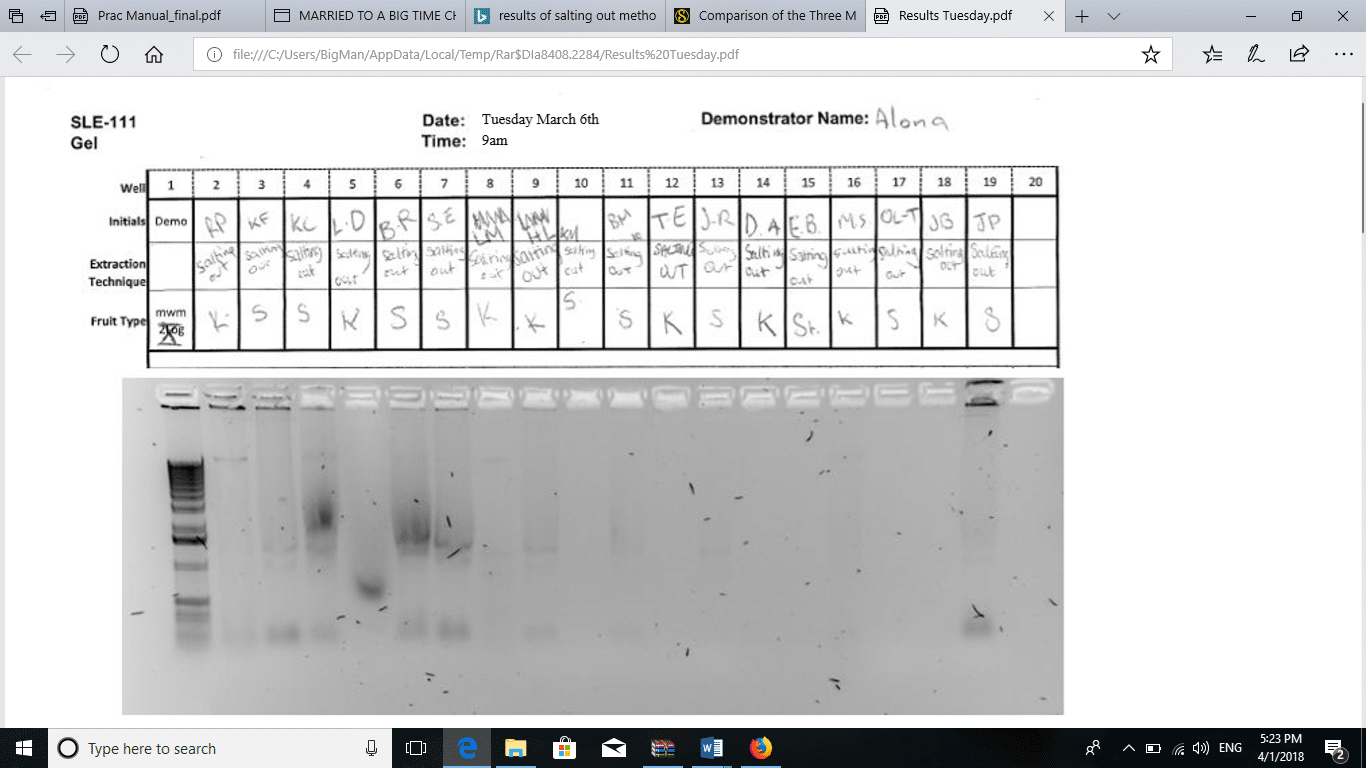
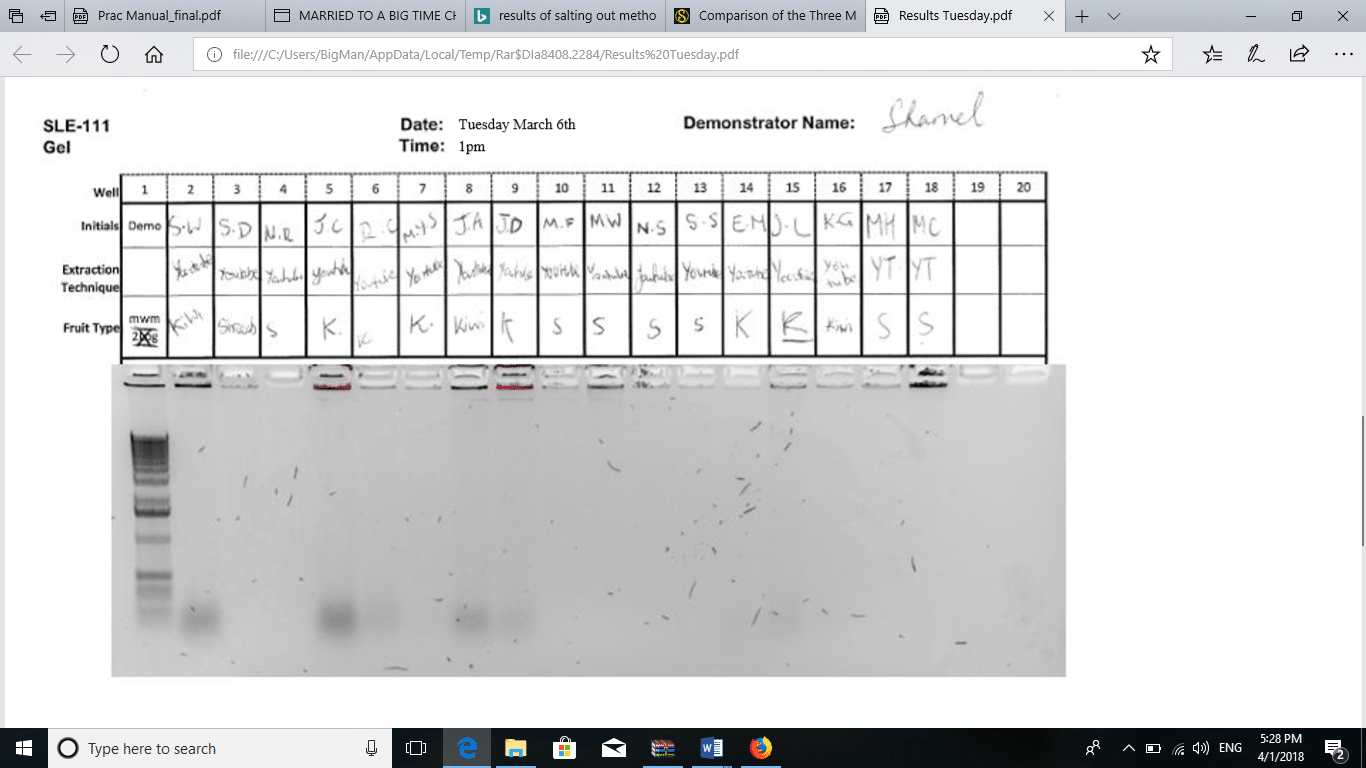
The addition of 170 µL of 5M NaCl to the sample precipitated all the proteins in the solution. The solution appeared murky. After the first centrifugation, two layers formed. The bottom layer contained solid protein particles, while the supernatant was a clear solution containing the dissolved DNA. After the addition of 100% ethanol to the supernatant, a white precipitate was formed. According to Chacon-Cortes and Griffiths (2014 p8), the addition of 100% ethanol causes DNA to precipitate, hence easy to separate from other components. The DNA pellets that formed after centrifugation were compared to the pellets formed from the YouTube method, the DNA pellet formed using the salting out method was larger (L), white, and opaque. The method of salting out is more effective than all the other methods of DNA extraction (Seeker et al., 2016, p135). Ultimately, the results of agarose gel electrophoresis are shown in Figure 1. Figure 2 shows the results obtained by the groups using the method on YouTube.
Figure 1: Gel electrophoresis for salting out method.
Figure 2: Gel electrophoresis for YouTube method.
Discussion of Results
The aim of the experiment was twofold; to extract DNA from kiwi and strawberry fruits and to compare the salting out method with the YouTube method. Based on the results of the experiments, all these aims were achieved. Adequate amounts of DNA were extracted from both kiwi fruit and strawberry fruits, demonstrating the validity of the salting out method. Moreover, the results obtained by the groups using the YouTube method were adequate for comparative analysis of the methods.
It was possible to remove the walls and membranes of the cells because proteinase K lysed the protein components of the cells, exposing all the other elements of the cells which include the DNA material in the nucleus. Proteinase K is an enzyme extracted from Engyodontium album and can digest even keratin and cellulose (Chacon-Cortes, and Griffiths, 2014 p6). It cleaves the proteins at the peptide bond next to the carboxyl group of amino acids which have blocked amino groups at the alpha position. The purpose of the TNES buffer was to provide favorable conditions for the optimum function of the enzyme. The contents of the buffer were 0.5% SDS and other components, which give an enzyme activity of approximately 120% (Chacon-Cortes, and Griffiths, 2014 p8). Additionally, the incubation at 37oC was optimum for protease activity. The addition of salt and ethanol to the solution forces the DNA to precipitate out. The ethanol dissolved all the non-polar substances, leaving the DNA in the aqueous phase.
During agarose gel electrophoresis, Seeker and colleagues (2016 p136) assert that there is inverse proportionality between the size of base pairs in the DNA and the distance moved through the gel. The assertion implies that DNA with short strands runs a longer distance along the gel while DNA with longer strands runs a shorter distance through the gel. Based on this argument, the YouTube method produced DNA with longer strands than the other method. Most of the DNA extracted by the YouTube method moved very short distances, while those obtained by salting out moved a considerable distance through the gel. The quantities of DNA obtained by salting out were comparable to those obtained using other methods like the phenol-chloroform extraction technique. The procedure has been widely used in forensics and laboratory studies.
The salting out technique has its limitations. The results obtained in this experiment are only applicable when the reagents and conditions are maintained. Any variation in the experimental conditions would lead to significant differences in the results. Additionally, it is quite tricky to amplify DNA extracted using the salting out method.
Conclusion
DNA extraction is the best discovery that ever graced the scientific world. Based on the numerous applications it has in diagnostics, treatment, genetic engineering, and forensics, DNA extraction is worth keen attention. Various techniques have been developed, as discussed at the onset of this report. Scientists are in constant search of a method that is efficient and cost-effective. Most researchers have proved to salt out a method to be profitable and straightforward to carry out. However, the method presents considerable difficulty when it comes to the amplification of the DNA. It also produces shorter DNA strands compared to other ways. Therefore, future studies should focus on either finding out the best method that will make amplification easier or finding a modification to salting out a method to minimize the afore-highlighted limitations.
Reference List
Tamari, F. and Hinkley, C.S., 2016. Extraction of DNA from plant tissue: review and protocols. In Sample Preparation Techniques for Soil, Plant, and Animal Samples (pp. 245-263). Humana Press, New York, NY.
Zhang, Y.H.P., Sun, J. and Ma, Y., 2017. Biomanufacturing: history and perspective. Journal of industrial microbiology & biotechnology, 44(4-5), pp.773-784.
Seeker, L.A., Holland, R., Underwood, S., Fairlie, J., Psifidi, A., Ilska, J.J., Bagnall, A., Whitelaw, B., Coffey, M., Banos, G. and Nussey, D.H., 2016. Method specific calibration corrects for DNA extraction method effects on relative telomere length measurements by quantitative PCR. PloS one, 11(10), p.e0164046.
Chacon-Cortes, D. and Griffiths, L.R., 2014. Methods for extracting genomic DNA from whole blood samples: current perspectives. Journal of Biorepository Science for Applied Medicine, 2014(2), pp.1-9.